Standardization of Culture Conditions
| Growing | Starvation | Conjugation | |
|---|---|---|---|
| Strains | CU428 | CU428 | CU428 X B2086 |
| Growing conditions | 1XSPP(1%PP), 30°C, 150 rpm shaking, 1/10 cell volume to flask size ( 2 L Erlenmeyer flasks for G and S, 1 L flasks for C); 200K cells/ml | ||
| Starvation conditions | 10 mM tris, pH 7.4-7.5, 100X antibiotic- antimycotic mix; without shaking; 200K cells/ml | ||
| Conjugation conditions | Mix cells starved separately for 18h | ||
| Time points | 100K, 350K and 1000K cells/ml (cell counter) | every 3 h from 0-15 h and 24 h (H1 mobility assay) | every 2 h from 0-18 h (observe the stages microscopically) |
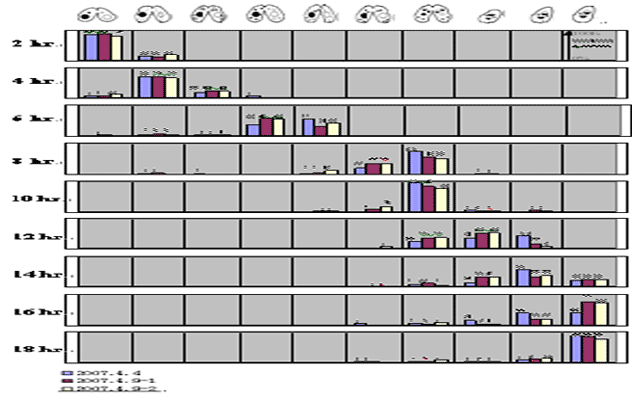
Stages During Conjugation
Method for Preparing RNA for Microarrays and RNA-Seq
(Jody Bowen and Wei Miao, 2/20/07 )
- 1. Used Qiashredder (Cat # 79654) and the RNeasy kit (cat. # 74624), from Qiagen. Contains RNA Protect Cell Reagent (Cat. # 76526) and RNeasy Plus Mini Kit (50 samples);
- Used normal wild-type CU428 strain obtained from the Bruns' lab at Cornell. Taken from our liquid nitrogen freezer stocks in January, 2007. Held as soybean stocks, which were used to start the first culture listed below.
- NOTE: Read the instructions in the RNeasy Kit since we have not included all the optional information or details. From pilot experiments, Jody got about 40 mg of RNA from 1 x 106 starved cells or 3.3 x 105 log cells. Those amounts of cells should, in theory, contain ~100 mg of RNA. Using 2X as many cells still gave RNA that looked good on a northern blot, but since we did not want to take a chance overloading the RNeasy system, we decided to use ~1.4 x 106 starved cells per sample. Following Jody's original test protocol, Wei Miao prepared three sets of starved cells. We have included the OD readings for the three starvation procedures that he has done. A northern was done and yielded the correct size for a large RNA message and was a nice sharp band, no degradation seen.
Starving Tetrahymena thermophila (CU428) Cells and isolating their RNA for Microarray Analysis
- First day, start cultures (2 days ahead of when they are needed for starvation): inoculate the cells into 25 ml 1X SPP (Super Proteose Peptone) in 125 ml flask, incubate at 30°C with shaking (~200 rpm);
- Second day, count the cells to make sure they are in log phase (2~4X105 cells/ml). Determine the amount of cells you will need to start cultures in order to have enough cells in log phase when you want to starve them. Then, inoculate the cells into 300 ml 1X SPP in a 2L flask, and incubate at 30°C with shaking (200 rpm). We assume ~2.5 hr doubling and started with 3125 cells/ml; it should take ~16h to reach ~ 200,000 cells/ml;
- Third day, count the cells to make sure they are in early log phase ( ~ 200,000
cells/ml). Determine the amount of cell culture you will need to collect to have
the required volume of starved cells at ~200,000/ml;
1). Spin cells down in sterile Corning 50ml plastic centrifuge tubes for 2 minutes at 1500 RPM in the Primo centrifuge (about 360 xg)
2). Pour off the medium as soon as the tubes stop spinning;
3). Resuspend the cells with ~15-20 ml 10 mM Tris buffer (pH 7.5), shake the tube gently to resuspend cells, combine into 2 tubes from the original 4 or 6 tubes;
4). Spin the cells down again at 1500 RPM X 2 minutes and pour off as much of the wash solution as possible;
5). Resuspend the cells in 10mM Tris and transfer them to a sterile 2L flask, and add sterile 10mM Tris to give a cell concentration of ~ 200,000 cells/ml. The liquid must be less than 1/10 of the total flask volume (200ml);
6). Add 2ml 100X antibiotic-antimycotic mix;
7). Incubate the starvation flask at 30°C without shaking - Remove cells for RNA isolation at 0, 3, 6, 9, 12, 15 and 24h. Count cells at the beginning and after last collection to be sure they are ~200,000cells/ml/li>
RNA Isolation
- For starved cells, at each time point collect 7ml cells ( ~ 200,000 cells/ml) using the sterile Corning 15 ml conical tubes: Spin the cells down at 1500 RPM (360 xg) X 2 minutes and immediately pour off as much of the supernatant as possible. The packed cell volume should be 150 μl or less
- Add 750 μl (5X volume of cells and residual supernatant) RNAprotect Cell Reagent to cells. Mix them well by pipetting. Transfer the mix into 1.5 ml RNase free microcentrifuge tube. Store at -20°C;
- Can store all samples at -20°C until it is convenient to continue with the RNA isolation (Qiagen says days or weeks). When ready to proceed, thaw all samples at room temperature. Centrifuge the cells in RNAprotect Cell Reagent for 5 min at 5000 xg. We used the Primo microfuge;
- Remove the supernatant completely by pipetting;
- Loosen the pellet by flicking the tube or vortexing;
- Add 350 μl Buffer RLT Plus to the sample (just before using, add 10 ul of beta mercaptoethanol per ml to the total amount of Buffer RLT Plus needed for the number of samples being processed). Dissolve the pellet completely by pipetting and vortexing (took ~3 min), and homogenize immediately as described below;
- Homogenization is done using the Qiashredder. Pipet the lysate directly into a QIAshredder spin column placed in a 2 ml collection tube, and centrifuge for 2 min at 16,000 xg, save the flow-through homogenate;
- Transfer the homogenate to a gDNA Eliminator spin column placed in a 2 ml collection tube. Centrifuge for 30s at 10,000 g. Discard the column, and save the flow-through;
- Add 350 μl 70% ethanol (made with DEPC-treated water), and mix thoroughly by vortexing. Do not centrifuge. Proceed immediately to next step;
- Transfer the 700 μl sample, including any precipitate, to an RNeasy Mini spin column placed in a 2 ml collection tube. Close the lid gently, and centrifuge for 15 s at 10,000g . Discard the flow-through;
- Add 700 μl Bffer RW1 to the RNeasy Mini spin column from step 10. Close the lid gently, and centrifuge for 15 s at 10,000g to wash the spin column membrane. Discard the flow-through;
- Add 500 μl Buffer RPE to the RNeasy Mini spin column. Close the lid gently, and centrifuge for 15 s at 10,000g . Discard the flow-through; Note: Buffer RPE is supplied as a concentrate. Be sure that ethanol is added to Buffer RPE before use
- Add 500 μl Buffer RPE to the RNeasy Mini spin column. Close the lid gently, and centrifuge for 2 min at 10,000g . Discard the flow-through;
- Place the RNeasy Mini spin column in a new 2 ml collection tube. Centrifuge at 10,000g for 1 min. Discard the flow-through;
- Place the RNeasy Mini spin column in a 1.5 ml collection tube. Add 50 μl RNase-free water directly to the spin column membrane. Close the lid gently, let sit ~ 1 min and centrifuge for 1 min at 10,000g to elute the RNA. Leave eluant with RNA in tube;
- Repeat step 15 with 30 μl RNase-free water;
- Store the RNA at -80 °C, To avoid thawing your samples later, remove 2 ul RNA into 0.5 ml RNase free tube to use for reading OD by NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE) and 4 ul for checking the quality by Bioanalyzer 1000 (Agilent, Palo Alto, CA).Remove cells for RNA isolation at 0, 3, 6, 9, 12, 15 and 24h. Count cells at the beginning and after last collection to be sure they are ~200,000cells/ml
Library construction and sequencing method for RNA-Seq
Poly-A mRNAs were isolated using Dynal magnetic beads (Invitrogen) and fragmented by heating at 94°C. First strand cDNAs were synthesized with reverse transcriptase and random hexamer primers, and then the second strands were synthesized with DNA polymerase and random hexamer primers. Double strand cDNAs were end-repaired and a single adenosine moiety was added. Illumina adapters were ligated and gel-electrophoresis was used to select DNA fragments between 200–250 bp in size. All the enzymes used above are included in the kit from Illumina. Libraries were PCR-amplified using Phusion polymerase. Sequencing libraries were denatured with sodium hydroxide and diluted in hybridization buffer for loading onto a single lane of an Illumina GA flowcell. Cluster formation, primer hybridization and pair-end sequencing were performed using proprietary reagents according to manufacturer-recommended protocols (https://icom.illumina.com/).




